The novel coronavirus (COVID-19) has become a worldwide pandemic that has caused massive disruptions at all levels of life, from government infrastructure and markets to our day-to-day interactions with each other. Like Zika, severe acute respiratory syndrome (SARS), and the H1N1 (swine flu) viruses before it, COVID-19 demands a dedicated and multi-pronged approach to stem its outbreak and to eventually defeat it. Containment and prevention through methods like social distancing and hand washing are techniques that can be practiced by everyone [1], but they require strict enforcement and diligence to be effective. Longer term solutions like vaccine development require significant investment and research to understand the mechanism by which the virus attacks the body and its cells.
Viruses replicate by entering cells and hijacking them to synthesize new viruses. Vaccines prevent this event from occurring by first exposing the body’s immune system to either an inactivated/weakened form of the virus or a key portion, such as its capsule or surface antigen. After this exposure, the body can then recognize the virus and neutralize it after a subsequent encounter. Determining which portion of the virus to inactivate or isolate to form a vaccine involves carefully studying its structure and discovering what binds to it in a sensitive and specific fashion.
Development of an accurate test to determine whether a person has been infected is a complementary step for ensuring a healthy populace. Virus detection methods include antibody tests, DNA or RNA tests, and antigen tests. Successful engineering of these tests involves understanding the components of the virus that can most specifically and sensitively identify it. For some viruses, this can involve looking for antibodies or viral antigens that a person has produced. In other cases, this may require extraction of the viral DNA or RNA to match a complementary strand [2]. Either strategy requires a high-content, highly sensitive technique that can accurately and reproducibly determine binding of these molecules.
Quartz crystal microbalance with dissipation (QCM-D) iis a real-time, label-free technique that has been used for several decades to accurately measure biomolecular binding. It utilizes a quartz sensor that oscillates at a resonant frequency when a voltage is applied. As mass is added to the sensor, the frequency of oscillation decreases, which is directly related to the mass added to the sensor. In addition to frequency changes, the technique also measures dissipation, which looks at how quickly the sensor comes to rest once the applied voltage is stopped. The speed at which this occurs is related to the rigidity of the mass attached. More rigid masses tend to come to rest slower compared to softer masses, since softer materials will tend to dissipate kinetic energy more readily. The technique has a sensitivity on the order of ng/cm2, which is suitable for measuring viral, DNA, and protein-protein interactions. Read more details on the the QCM-D technique on our techniques page.
Supported Lipid Bilayers as a Model System for Virus Interactions
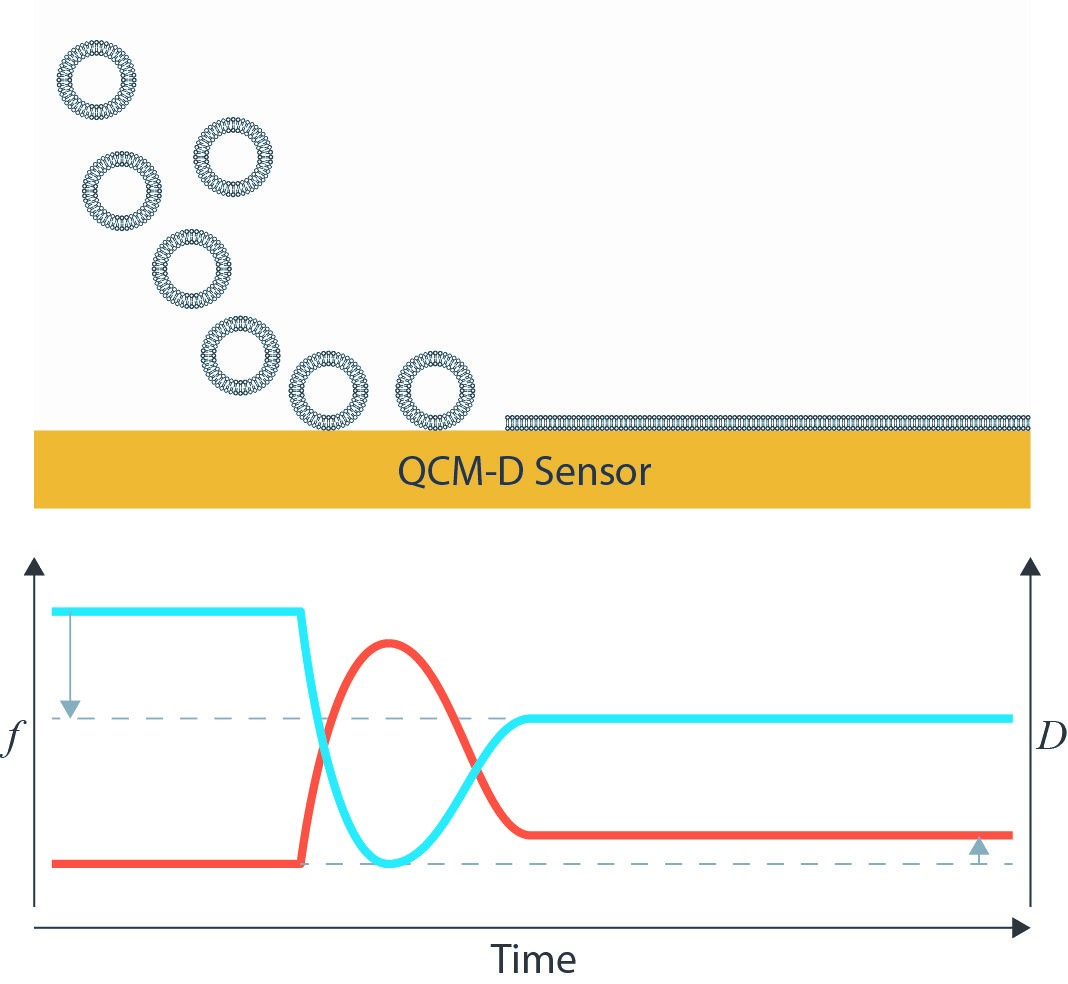
The applicability of the QCM-D platform as a sensor for biomolecular interactions is well-established. After first adsorbing or immobilizing a receptor molecule on a surface, its interactions with a target ligand can then be monitored. A common substrate for QCM-D studies is a supported lipid bilayer because it serves as a mimic for a cell membrane. These lipid bilayers are often first synthesized as a lipid vesicle. The gravimetric nature of the QCM-D technique is ideal for tracking the lipid bilayer formation from its vesicle form. Depending on surface chemistry, it may be possible for these vesicles to adsorb intact or to rupture and form the intended lipid bilayer [3]. A clear frequency decrease from the initial adsorption and subsequent frequency increase due to the loss of coupled solvent from the interior of the vesicle can be tracked. Dissipation initially increases due to the adsorption of a soft, hydrated mass, and then decreases as the vesicle unfolds into a rigid bilayer.
Researchers can leverage these bilayers by inserting proteins that interact specifically with a virus protein or shell. Cho et al. looked at how the NS5A protein from hepatitis C bound to a Huh7 (liver cell)-derived membrane [4]. They hypothesized that the amphipathic helix (AH) binding domain in the NS5A protein was responsible for the binding between the liver cell and the hepatitis C protein. Using QCM-D, they observed that the NS5A protein adsorbed to the membrane, and that when the AH domain was replaced with an altered form, the binding no longer occurred. Additionally, when the Huh7 membrane was exposed to trypsin, which cleaves proteins from the surface, the NS5A protein also no longer bound. This indicates the specificity of the AH binding site to certain surface proteins on the liver cell surface. Several years later, the group was able to narrow down the interaction between NS5A and the liver cell to phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), which interacts with a BAAPP domain in the NS5A protein. These results were again confirmed by comparing QCM-D responses of the PI(4,5)P2 with variants and inhibitors [5].
Protein Immobilization through Surface Chemistry
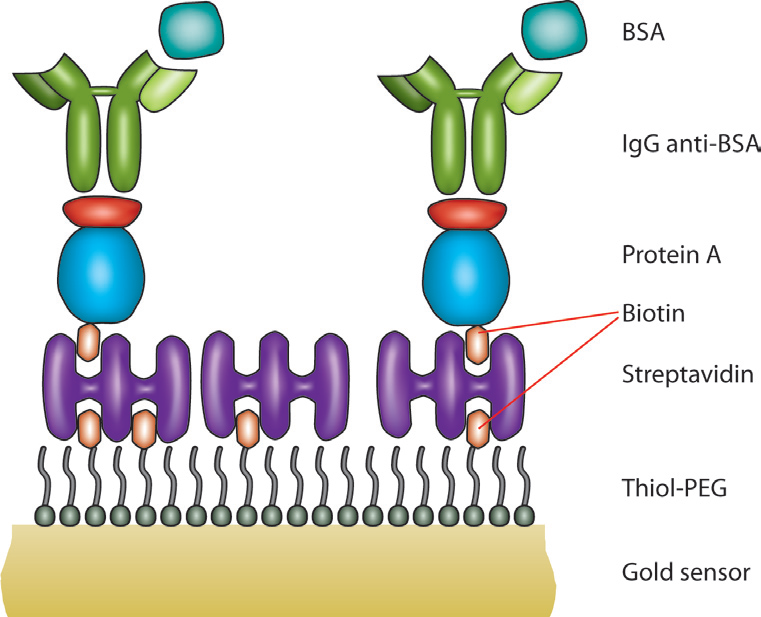
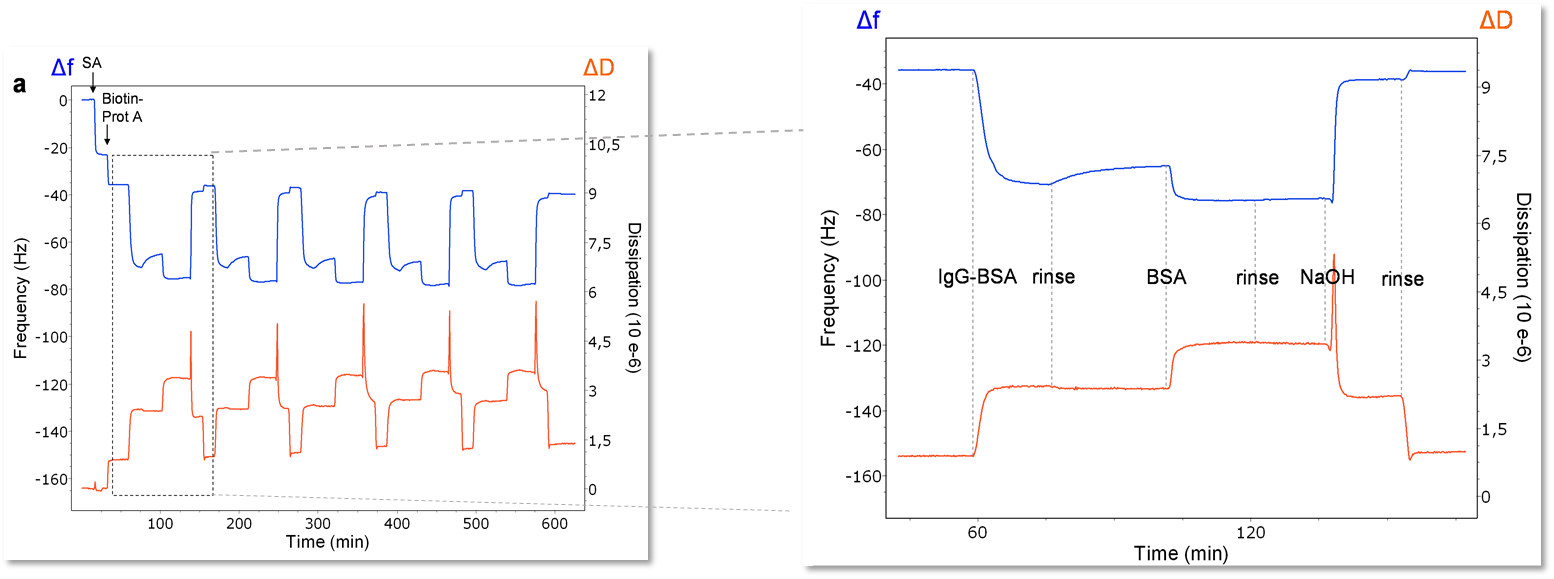
One other primary advantage of QCM-D is that due to its gravimetric nature, any surface that can be coated onto the quartz sensor can be used as a substrate. This allows for a wide application space including topics such as electrochemistry, fouling, drug packaging interactions, and much more. For viral studies, surface chemistry techniques can be used to make a biosensor-type surface as seen in Figure 2. A thiolated poly(ethylene glycol) (PEG) surface can be adsorbed with a biotin-streptavidin layer, allowing for a “layer-by-layer” buildup of other proteins to eventually immobilize an antibody of interest. This antibody can then serve as a detector for a specific molecule, in this case, bovine serum album (BSA). The entire interaction can be monitored in situ with the QCM-D to ensure that it proceeds as expected. Additionally, the surface can be regenerated and re-built as seen in Figure 3. On the left side of Figure 3, five regeneration steps are performed, each of which reproducibly returns the frequency and dissipation values to a stable baseline. The right side of Figure 3 shows in more detail the steps in each assay and regeneration cycle, where the BSA is clearly seen interacting with the anti-BSA antibody. NaOH is then flowed over the sensor surface to remove the anti-BSA and BSA from the surface. A rinsing step is performed using the base buffer between each step to remove loosely bound molecules.
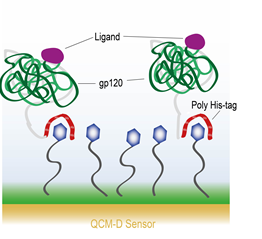
Surface chemistry to covalently attach molecules to a surface is also possible. Lee et al. describe a process where they covalently bind 3,3′dithiobis[N-(5-amino-5-(carboxypentyl) propionamide)-N,N′-diacetic acid]dihyrochloride (NTA) to a gold surface. The NTA then interacts with HIV-1 envelope protein gp120 via a hexahistidine tag (his-tag) on the protein [6]. A schematic of the experimental setup is shown in Figure 4.. They then exposed the gp120 to four different ligand molecules – sCD4, HNG-156, BMS-806, and NBD-556. With every ligand molecule except BMS-806, they observed a conformational change in the gp120 protein such that the gp120 appeared to contract upon coupling with the ligand. This is observed through a decrease in the dissipation and an increase in the frequency, indicating a mass loss. They mention that the QCM-D response is consistent with the protein simultaneously collapsing and ejecting coupled water from the system. The BMS-606 molecule, on the other hand, showed only a temporary interaction with the gp120 and was rinsed off.
DNA Hybridization
Several QCM-D studies have examined DNA hybridization on the quartz sensor. Gunnarsson et al. looked at binding specificity of a 15 mer DNA immobilized to through a biotinylated PLL-g-PEG molecule to a 30 mer complementary strand attached to a lipid vesicle [7]. They then generated a single base pair mismatch on the 30 mer strand and observed roughly 60% fewer vesicles bound to the surface.
Rawle et al. examined hybridization using a slightly different method. Instead of using a biotinylated capture surface to immobilize the DNA, they used a positively charged poly(ethyleneimine) (PEI) electrolyte layer as a charged surface to allow the DNA to electrostatically bind to the surface [8]. Upon adsorbing a solution of denatured DNA, they observed an initial sharp frequency shift, and then a second, slower shift that indicated a secondary adsorption step. They attributed this slower shift to hybridization of the DNA strands adsorbed on the surface of the sensor.
Concluding Remarks
Because of its sensitivity to small molecules, flexible sensor configurations, and real-time, label-free sensing, QCM-D is an excellent choice for researchers looking to expand their capabilities to closely examine mechanistic interactions between viruses and viral particles and their target molecules. Using this technique, information about specific binding properties can be determined. Additionally, biophysical details about the protein or molecular conformation can also be discerned.
References
[1] Center for Disease Control and Prevention, https://www.cdc.gov/coronavirus/2019-ncov/prepare/prevention.html, 2020.
[2] World Health Organization, “Vaccine Safety Basics,” https://vaccine-safety-training.org/home.html, 2020.
[3] C. . Keller and B. H. Kasemo, “Surface Specific Kinetics of Lipid Vesicle Adsorption Measured with a Quartz Crystal Microbalance,” Biophysical Journal, vol. 75, no. 3, pp. 1397-1402, 1998.
[4] N.-J. . Cho, K. H. Cheong, C. . Lee, C. W. Frank and J. S. Glenn, “Binding Dynamics of Hepatitis C Virus’ NS5A Amphipathic Peptide to Cell and Model Membranes,” Journal of Virology, vol. 81, no. 12, pp. 6682-6689, 2007.
[5] N.-J. . Cho, C. . Lee, P. S. Pang, E. A. Pham, B. . Fram, K. . Nguyen, A. . Xiong, E. H. Sklan, M. . Elazar, E. S. Koytak, C. . Kersten, K. K. Kanazawa, C. W. Frank, J. S. Glenn and J. S. Glenn, “Phosphatidylinositol 4,5-Bisphosphate Is an HCV NS5A Ligand and Mediates Replication of the Viral Genome,” Gastroenterology, vol. 148, no. 3, pp. 616-625, 2015.
[6] H.-S. . Lee, M. . Contarino, M. . Umashankara, A. . Schön, E. I. Freire, A. B. Smith, I. M. Chaiken and L. S. Penn, “Use of the quartz crystal microbalance to monitor ligand-induced conformational rearrangements in HIV-1 envelope protein gp120,” Analytical and Bioanalytical Chemistry, vol. 396, no. 3, pp. 1143-1152, 2010.
[7] A. Gunnarsson, P. Jonsson, R. Marie, J. O. Tegenfeldt and F. Hook, “Single-Molecule Detection and Mismatch Discrimination of Unlabeled DNA Targets,” Nano Letters, vol. 8, no. 1, pp. 183-188, 2008.
[8] R. J. Rawle, C. D. Selassie and M. S. Johal, “Creation of mammalian single- and double-stranded DNA surfaces: a real-time QCM-D study.,” Langmuir, vol. 23, no. 19, pp. 9563-9566, 2007.
